Feature matching
Feature matching is a critical part in many vision problems, such as estimation of various matrices (projection, homography, essential, fundamental, relative post, etc), tracking, SfM and SLAM, just to name a few. I will discuss here in this post implementation of the feature matching algorithms implemented in open3DCV.
Result

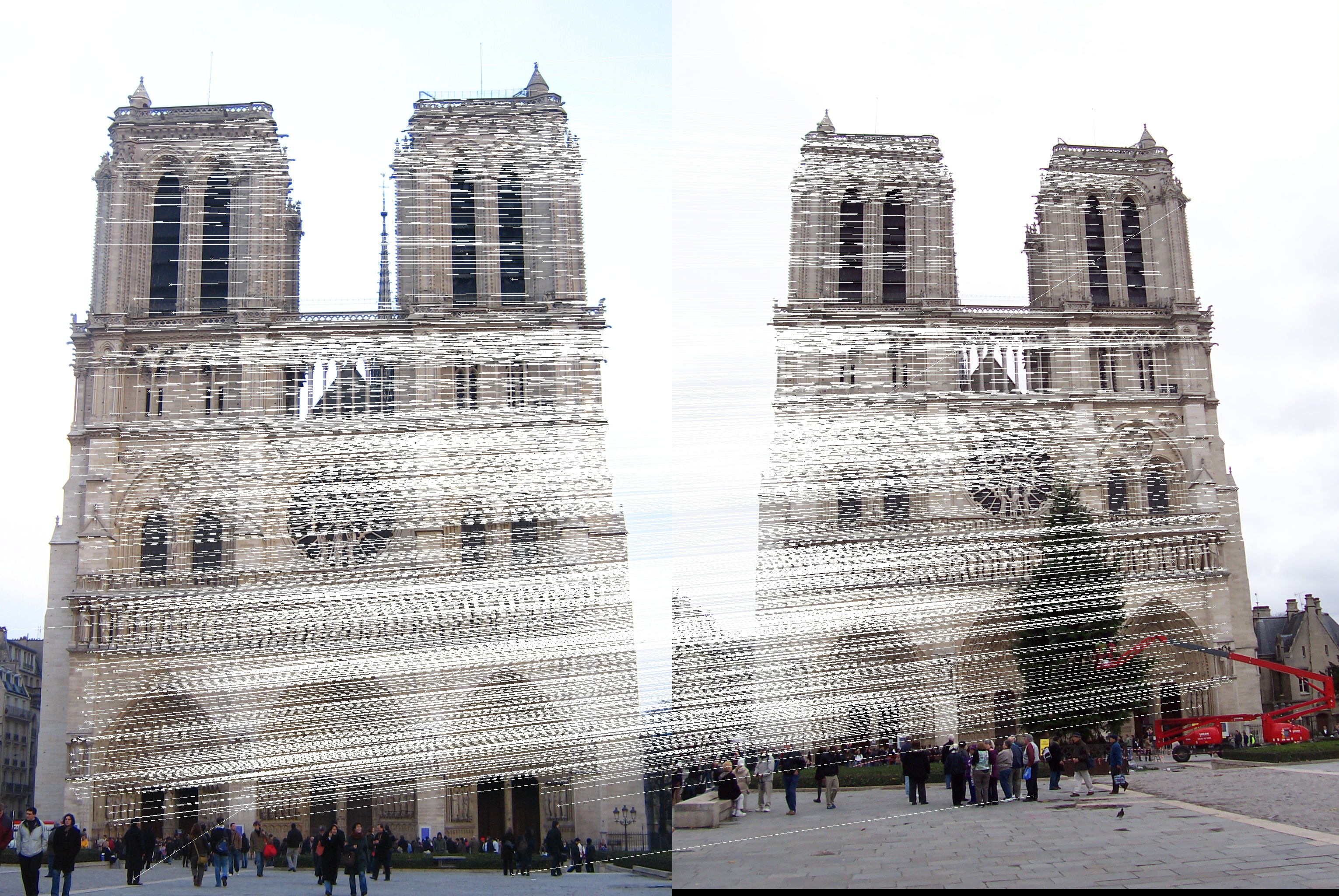
Base class: matcher
There is a base class matcher that all actual matching algorithms need to extend. The match method takes two vectors of descriptors and returns the indexes of the matching pair. Note that there are two declarations of match method, the second provides an additional parameter - a function pointer, that allows to use the user-defined distance function.
class Matcher
{
public:
Matcher() { };
Matcher(Matcher_Param r_matcher_param);
virtual ~Matcher() { };
virtual void init_param(Matcher_Param r_matcher_param) = 0;
virtual int match(const vector<Vecf>& desc1, const vector<Vecf>& desc2, vector<Match>& matches) = 0;
virtual int match(const vector<Vecf>& desc1, const vector<Vecf>& desc2, vector<Match>& matches, float (*dist_metric)(const Vecf& desc1, const Vecf& desc2)) = 0;
protected:
Matcher_Param matcher_param_;
};
Parameter: Matcher_Param
We need a structure type to pass the parameters to the specific matcher class, which is defined as follows
struct Matcher_Param
{
// all matchers
float ratio = 0.8;
// Flann matcher
int ndims = 128;
int leaf_max_size = 10;
int nresults = 3;
Matcher_Param() { };
Matcher_Param(const float r_ratio) : ratio(r_ratio) { };
Matcher_Param(const float r_ratio, const int r_ndims, const int r_leaf_max_size, const int r_nresults) :
ratio(r_ratio), ndims(r_ndims), leaf_max_size(r_leaf_max_size), nresults(r_nresults) { };
};
Data structure: Match
This is the class to store the matching feature points, I should probably rename it to DMatch to avoid confusion.
class Match
{
public:
Match () { };
Match (int r_ikey1, int r_ikey2, float dist) :
ikey1_(r_ikey1), ikey2_(r_ikey2), dist_(dist) { };
int ikey1_;
int ikey2_;
float dist_;
};
Distance metrics
I implemented two of the most used distance metrics for now, sum of squared distance (SSD) and sum of absolute difference (SAD). The can be passed onto the function pointer when call the matcher method.
inline float l2_dist(const Vecf& desc1, const Vecf& desc2)
{
// return (desc1 - desc2).squaredNorm();
return (desc1 - desc2).norm();
};
inline float l1_dist(const Vecf& desc1, const Vecf& desc2)
{
return (desc1 - desc2).lpNorm<1>();
}
Brute-force method
This is the simplest, yet most computationally expensive method. It iterates all possible point pairs and see if the cost function is below a user-specified threshold.
inline int Matcher_Brute_Force::match(const vector<Vecf>& desc1, const vector<Vecf>& desc2, vector<Match>& matches)
{
return match(desc1, desc2, matches, l2_dist);
}
inline int Matcher_Brute_Force::match(const vector<Vecf>& desc1, const vector<Vecf>& desc2, vector<Match>& matches, float (*dist_metric)(const Vecf& desc1, const Vecf& desc2))
{
float dist = 0, min_dist = 1e8, sec_min_dist = 1e8, ratio = matcher_param_.ratio;
int ind_min_key = 0;
for (int i = 0; i < desc1.size(); ++i)
{
min_dist = sec_min_dist = 1e8;
for (int j = 0; j < desc2.size(); ++j)
{
dist = dist_metric(desc1[i], desc2[j]);
if (dist < min_dist)
{
sec_min_dist = min_dist;
min_dist = dist;
ind_min_key = j;
}
else if (dist < sec_min_dist)
{
sec_min_dist = dist;
}
}
if (min_dist < ratio * sec_min_dist)
{
Match m(i, ind_min_key, min_dist);
matches.push_back(m);
}
}
return 0;
}
ANN method
FLANN is short for Fast Library for Approximate Nearest Neighbors, which is proposed by Marius Muja and David Lowe. There are some similar libraries, such as ANN, ANNOY. Here is a benchmark for comparing the performance of approximate nearest neighbors algorithms. I used a simplified version of FLANN called nanoflann since I want the dependencies of open3DCV as simple as possible.
My implementation is adapted from the example code vector_of_vectors_example.cpp. There is one place that needs a heads-up, this function call of init(IndexType* indices_, DistanceType* dist_) has to be invoked everytime you do a search of nearest neighbor. The reason is best explained using the source code, see below
// implementation from nanoflann.h
inline void init(IndexType* indices_, DistanceType* dists_)
{
indices = indices_;
dists = dists_;
count = 0;
if (capacity)
dists[capacity-1] = (std::numeric_limits<DistanceType>::max)();
}
As we can see that init need to re-initialize dist[] to max values. Otherwise, the distances from the previous search is still stored in dist[]. The full implementation is as follows:
inline int Matcher_Flann::match(const vector<Vecf>& desc1, const vector<Vecf>& desc2, vector<Match>& matches)
{
return match(desc1, desc2, matches, l2_dist);
}
inline int Matcher_Flann::match(const vector<Vecf>& desc1, const vector<Vecf>& desc2, vector<Match>& matches, float (*dist_metric)(const Vecf& desc1, const Vecf& desc2))
{
// use the one with larger keypoints to construct kd-tree
size_t nkeys1 = desc1.size(), nkeys2 = desc2.size();
const vector<Vecf>& desc_source = nkeys1 < nkeys2 ? desc1 : desc2;
const vector<Vecf>& desc_target = nkeys1 < nkeys2 ? desc2 : desc1;
Matching_Dirc matching_dirc = nkeys1 < nkeys2 ? FORWARD : BACKWARD;
// construct a kd-tree index
typedef KDTreeVectorOfVectorsAdaptor<vector<Vecf>, float> kd_tree_t;
int leaf_max_size = matcher_param_.leaf_max_size;
kd_tree_t desc_index(matcher_param_.ndims, desc_target, leaf_max_size /* max leaf */);
desc_index.index->buildIndex();
// do knn searches
int nresults = matcher_param_.nresults;
vector<size_t> ret_indexes(nresults);
vector<float> dists(nresults);
nanoflann::KNNResultSet<float> result_set(nresults);
for (int i = 0; i < desc_source.size(); ++i)
{
result_set.init(&ret_indexes[0], &dists[0]); // has to be inside loop, there is a 'dist' variable that needs to be set as MAX
desc_index.index->findNeighbors(result_set, desc_source[i].data(), nanoflann::SearchParams(10));
if (dists[0] < matcher_param_.ratio * dists[1])
{
if (matching_dirc == FORWARD)
{
Match match(i, static_cast<int>(ret_indexes[0]), dists[0]);
matches.push_back(match);
}
else
{
Match match(static_cast<int>(ret_indexes[0]), i, dists[0]);
matches.push_back(match);
}
}
}
return 0;
}